
Multiple chromosomal inversions contribute to adaptive divergence of a dune sunflower ecotype - Huang - 2020 - Molecular Ecology - Wiley Online Library

Discovery of a polymorphism in 'CA60' that is unique to muRdr1A gene in... | Download Scientific Diagram
Linkage mapping and QTL analysis of flowering time using ddRAD sequencing with genotype error correction in Brassica napus
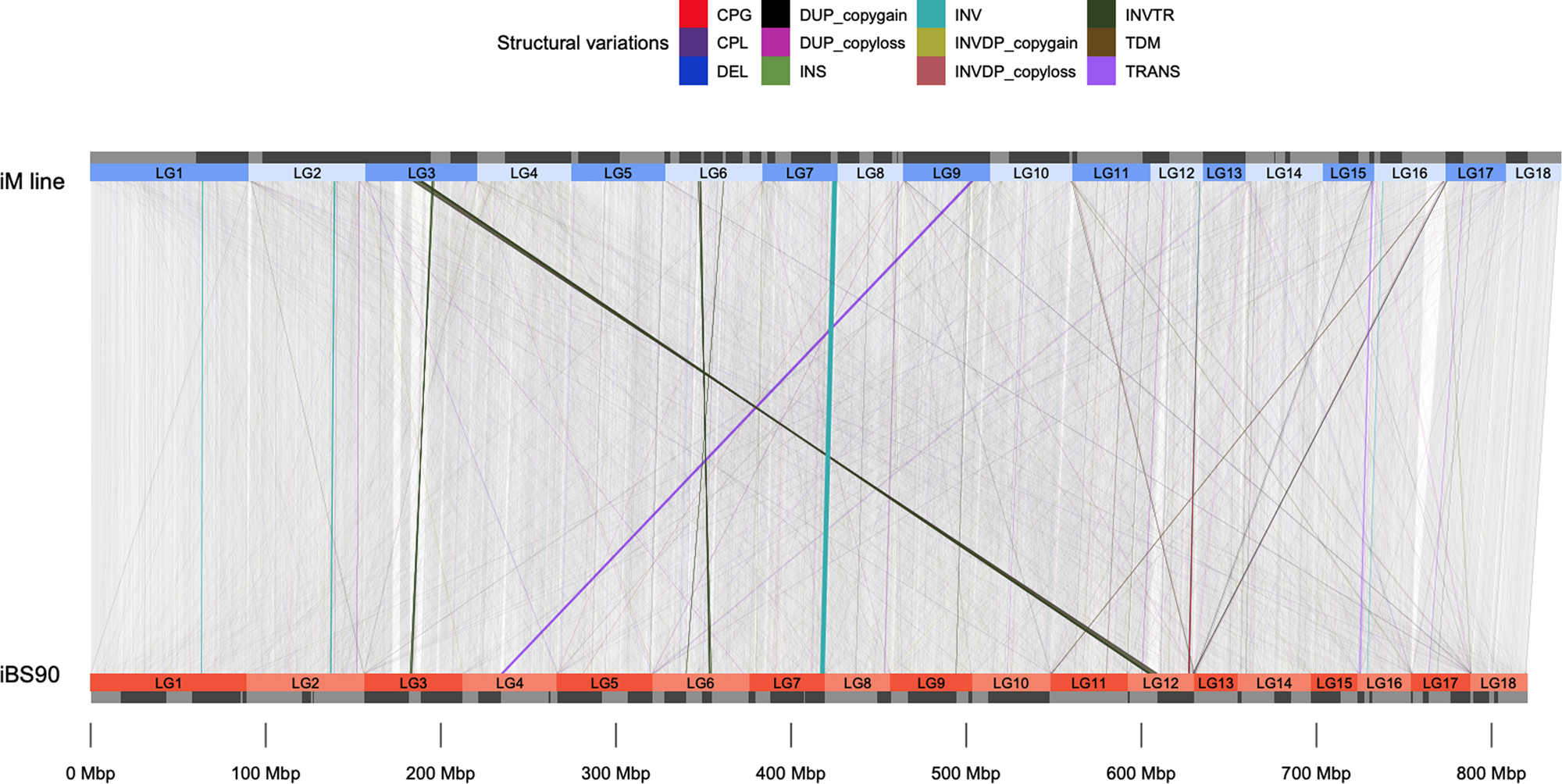
Compatibility between snails and schistosomes: insights from new genetic resources, comparative genomics, and genetic mapping | Communications Biology

Plants | Free Full-Text | Relocation of Sr48 to Chromosome 2D Using an Alternative Mapping Population and Development of a Closely Linked Marker Using Diverse Molecular Technologies

Plants | Free Full-Text | Relocation of Sr48 to Chromosome 2D Using an Alternative Mapping Population and Development of a Closely Linked Marker Using Diverse Molecular Technologies
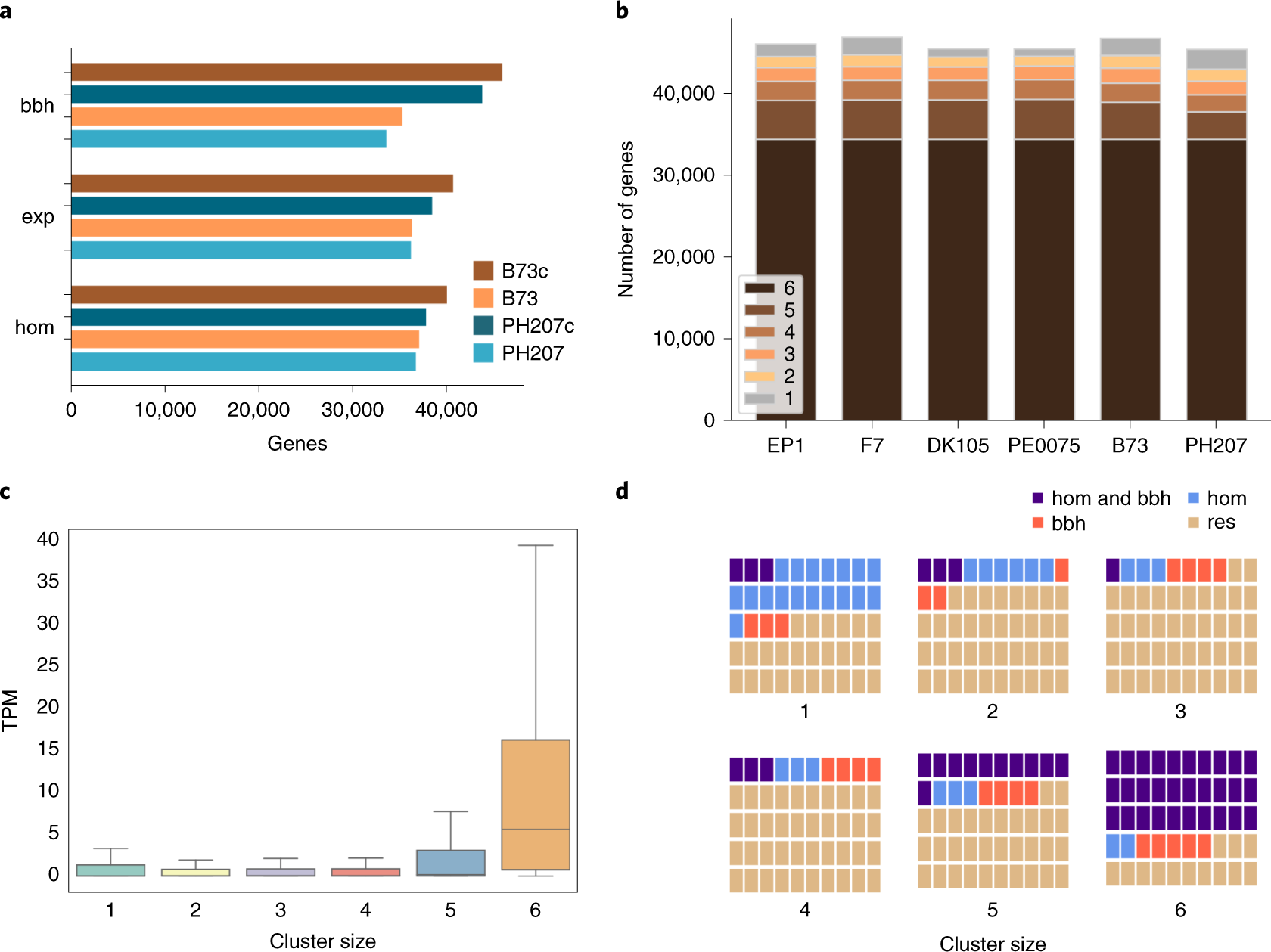
European maize genomes highlight intraspecies variation in repeat and gene content | Nature Genetics

Linkage mapping and QTL analysis of flowering time using ddRAD sequencing with genotype error correction in Brassica napus | bioRxiv

Multiple chromosomal inversions contribute to adaptive divergence of a dune sunflower ecotype - Huang - 2020 - Molecular Ecology - Wiley Online Library

Plants | Free Full-Text | Relocation of Sr48 to Chromosome 2D Using an Alternative Mapping Population and Development of a Closely Linked Marker Using Diverse Molecular Technologies

Genomic and Molecular Characterization of Wheat Streak Mosaic Virus Resistance Locus 2 (Wsm2) in Common Wheat (Triticum aestivum L.) - Frontiers

Plants | Free Full-Text | Relocation of Sr48 to Chromosome 2D Using an Alternative Mapping Population and Development of a Closely Linked Marker Using Diverse Molecular Technologies

Plants | Free Full-Text | Relocation of Sr48 to Chromosome 2D Using an Alternative Mapping Population and Development of a Closely Linked Marker Using Diverse Molecular Technologies

Natural Selection Shapes Variation in Genome-wide Recombination Rate in Drosophila pseudoobscura - ScienceDirect

Natural Selection Shapes Variation in Genome-wide Recombination Rate in Drosophila pseudoobscura - ScienceDirect

PDF) Relocation of Sr48 to Chromosome 2D Using an Alternative Mapping Population and Development of a Closely Linked Marker Using Diverse Molecular Technologies

Improving Nelumbo nucifera genome assemblies using high‐resolution genetic maps and BioNano genome mapping reveals ancient chromosome rearrangements - Gui - 2018 - The Plant Journal - Wiley Online Library
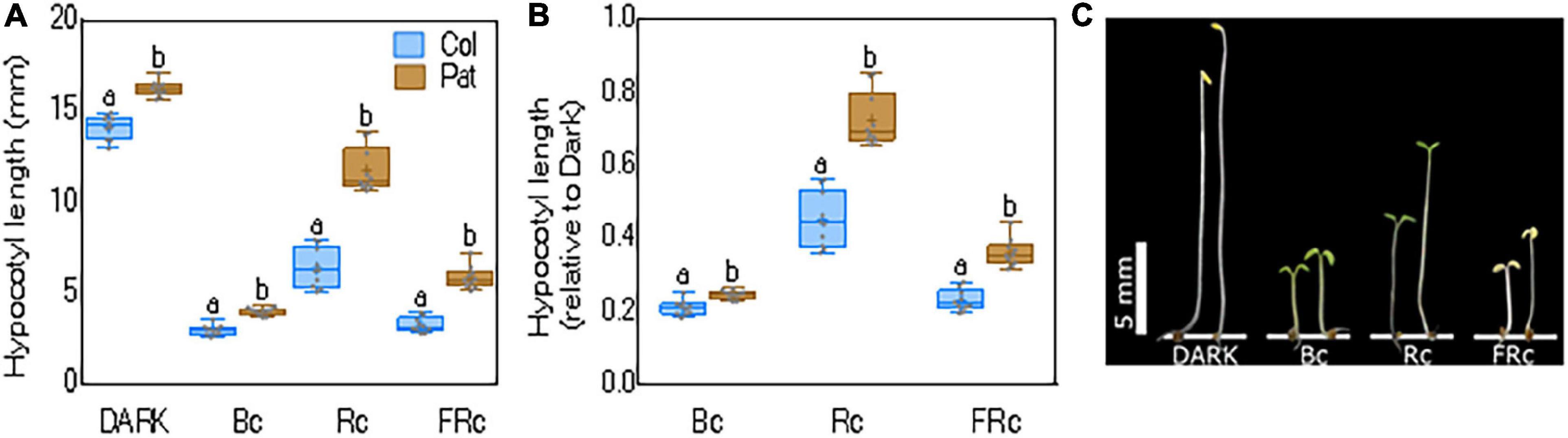
Frontiers | Ultra-High-Density QTL Marker Mapping for Seedling Photomorphogenesis Mediating Arabidopsis Establishment in Southern Patagonia


